Jeff Lichtman in front of his garden and home in Cambridge, MA (Courtesy of Daniel Berger)
Neuroscientist Jeff Lichtman on Being Humbled by Nature
by Parizad Bilimoria
From drops of pond water to slivers of brain tissue, Jeff Lichtman has always enjoyed looking inside things. His interest in the microscopic realm began back when he was a young boy growing up in Westchester, New York. The son of a hematologist, he had a small research grade microscope in his bedroom from second grade on, complete with its own oil immersion lens.
“I kind of took it for granted. It didn’t even occur to me that this was bizarre,” he says.
One of his favorite pastimes was studying the universe of microorganisms dwelling in local pond water. “I had a plastic Tupperware thing with leaves and water,” he recalls with a smile. “It smelled horrible.”
Life’s not all that different now that Lichtman is a world-renowned neurobiologist and faculty member at Harvard, recently elected to the National Academy of Sciences. Microscopes are still his favorite toys. His current fascination is a 61 beam scanning electron microscope custom-built for his lab, which came all the way from the Zeiss headquarters in Germany alongside an entourage of technicians. And while nowadays he uses his microscopes mainly to look at brains, Lichtman still loves his leaves and water. He and his wife are serious gardeners whose Cambridge, MA backyard drew in hundreds of visitors during a recent “Secret Gardens of Cambridge” tour.
The latest scientific passion for Lichtman, Jeremy R. Knowles Professor of Molecular and Cellular Biology and Ramón y Cajal Professor of Arts and Sciences in the Department of Molecular and Cellular Biology, and a member of the Center for Brain Science and Conte Center at Harvard, is mapping the “connectome”—or complete wiring diagram—of the mammalian brain. This is a project that his lab and others have been developing technology for over the past decade or more, and is expected to be significantly more challenging than the Human Genome Project—whose goal, ambitious back in the 1990s, was to determine the DNA sequence of all the genes in a human being.
The connectome needs to be mapped at many different spatial and temporal scales and forms an undeniably more complex dataset. For starters, the cerebral cortex in adult humans is thought to contain over 160 trillion sites of communication between neurons, called synapses. Properly identifying these synapses requires imaging at a resolution finer than the wavelength of light, and following tangles of very thin cables extending from neurons, called axons, for long distances—through extremely dense three dimensional terrain that must be analyzed piece-by-piece and then reconstructed computationally.
Learning by looking
So what drives this crazy quest? Why devote one’s life to such difficult work?
“I really love looking at data,” Lichtman explains. “The field we’re in in this lab is focused on seeing things that haven’t been seen before… we don’t actually know what we’re looking for until we see it.” He likens the excitement of seeing something new in the brain to how an astronomer feels looking at Hubble telescope images or how early explorers must have felt when coming across land masses not on their map.
“That for me is the favorite part of this job,” he declares. “This sense of discovery, of revelation—as opposed to ‘Aha, I knew I was right.’”
The Holy Grail for Lichtman has always been to learn from observing the natural world rather than manipulating it. He’s a tireless advocate for the kind of agnostic, question-driven science that uses data to generate hypotheses more than hypotheses to generate data.
Lichtman’s love of discovery can be traced back to his first findings in graduate school at Washington University in St. Louis. The first PhD student of neuroscientist Dale Purves, now at Duke University, Lichtman started his career studying neurons that control saliva secretion in rats. Certain clusters of these neurons receive inputs from the brainstem, and Lichtman noticed that these inputs were much more numerous in baby rats than adults. As the pups grew older, unnecessary neural connections appeared to be pruned away.
“I remember thinking that this has probably been true for millions of years,” he recalls, “but I am certainly the first human being to have noticed this.”
“That’s what science is about,” he adds, laughing about his level of enthusiasm for neurons that innervate rodent salivary glands. “Seeing something that hasn’t been seen before.”
How neural connectivity changes as babies grow up has continued to intrigue Lichtman throughout his career. A trailblazer in the interrelated fields of developmental neurobiology and biological imaging, Lichtman eventually followed up the salivary gland work with extensive studies of synapse elimination at the neuromuscular junction—the peripheral nervous system structure where neurons connect with the muscles they power. There too, he and colleagues observed a pruning in the neural inputs to muscles that occurs early in life, as animals explore their environments. These classic studies, requiring painstaking reconstructions of neural inputs onto individual muscle fibers followed over a series of time points and carried out in full molecular glory, are now a mainstay of neurobiology textbooks.
Ways to see more
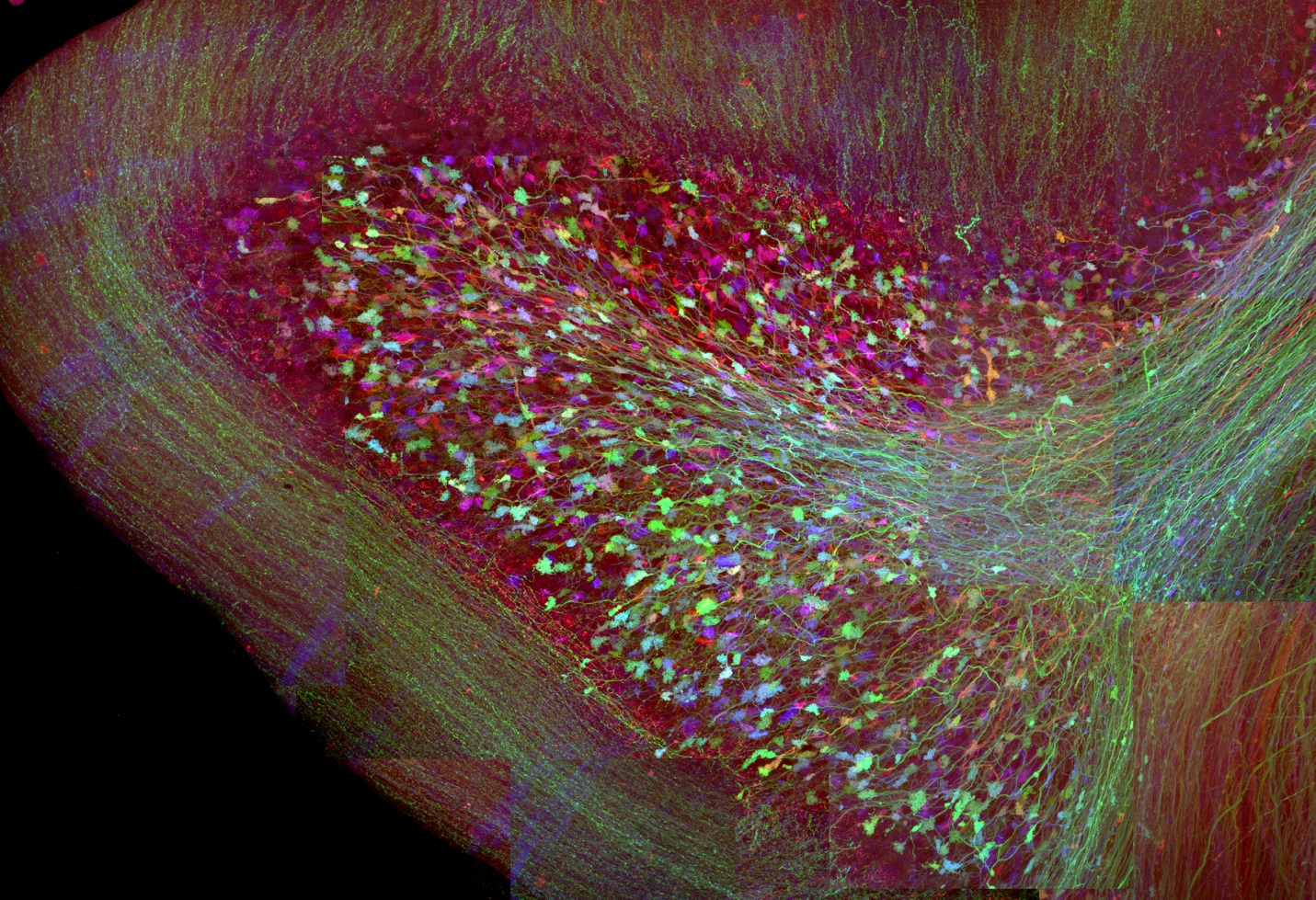
Brainbow in the Mouse Cerebellum (Image from 2007 study reported in Nature, featuring the work of Jean Livet, Tamily A. Weissman, Hyuno Kang, Ryan W. Draft, Ju Lu, Robyn A. Bennis, Joshua R. Sanes & Jeff W. Lichtman.)
Brainbow in the Mouse Cerebellum (Image from 2007 study reported in Nature, featuring the work of Jean Livet, Tamily A. Weissman, Hyuno Kang, Ryan W. Draft, Ju Lu, Robyn A. Bennis, Joshua R. Sanes & Jeff W. Lichtman.)
Never one to linger in a comfort zone, in more recent years Lichtman has started asking questions about synapse development in the central nervous system, between neurons instead of neurons and muscles. He’s also brought together teams of experts in imaging, genetics, histology, automation and computer science to develop ways to see more of the nervous system.
To begin to fathom how the brain works, Lichtman reasons that we need to see what’s inside at high resolution. That’s why he’s so excited about connectomics. To probe the basis of learning and memory, he argues that we need to see how the brain changes as we develop from infants into children and adults—following across development not just a few cherry-picked cells, but the full heterogeneous mass of cellular structures packed into a chunk of brain tissue.
The same goes for probing the basis of psychiatric disorders such as autism or schizophrenia, which he and others suspect may be “connectopathies”—or disorders of neural circuit development. How can we know what’s wrong in those disorders, he asks, if we lack basic stats about connectivity in a “normal” brain?
Towards these goals, Lichtman has explored both light and electron microscopy-based avenues for capturing more of the complexities of the nervous system. In collaboration with Joshua Sanes, also a faculty member in the Department of Molecular and Cellular Biology at Harvard and director of the Center for Brain Science, Lichtman co-led a team that invented “Brainbow”—a genetics trick in which the wires of the nervous system are lit up in a multitude of colors using different blends of fluorescent proteins. Just like a long tangle of computer cables is easier to work with when color-coded, this strategy allows researchers to distinguish and follow for long distances very densely packed, intertwined sets of axons.
Brainbow enables morphological analyses that before would not have been possible with fluorescence microscopy. Since its debut in transgenic mice in 2007, the world of Brainbow has blossomed to include cells outside the nervous system, organisms such as flies and fish, novel methods of gene delivery, as well as several new versions of the original mouse lines.
A Tiny Piece of the Connectome: A small portion of the apical dendrite of a pyramidal neuron in the mouse cortex is shown in red. The multicolor cylinder reveals a thin sliver of the surrounding brain tissue. Reconstruction based on serial scanning electron microscopy. (This image represents the work of Daniel Berger (formerly at MIT), Bobby Kasthuri and Richard Schalek in the lab of Jeff Lichtman.)
At the same time Lichtman’s lab is making major improvements to the methods for large scale electron microscopy analyses of the brain. Electron microscopy allows visualization of structures smaller than the wavelength of light—making it possible to clearly see the organelles within neurons, such as the mitochondria that give them their energy or the neurotransmitter vesicles whose release at synapses allows neurons to talk to one another. And while standard fluorescence microscopy can be used to predict synaptic sites, it’s the neurotransmitter vesicles revealed by electron microscopy that may tell how powerful synapses are.
While these virtues have long been evident, and it has for some time been possible to image ultrathin slices of brain tissue by electron microscopy and piece together three dimensional reconstructions, it’s an incredibly slow process that has until recently allowed only very small, fragmented glimpses into connectome. But thanks to the recent acceleration of several steps in the pipeline from brain slicing and imaging to analysis, it’s now possible to reconstruct in one thousandth the time it would have taken ten years ago. These improvements are allowing Lichtman’s team to gain increasingly larger glimpses into the connectome and imagine a day when it will be possible to attain synapse-level reconstructions of entire brains.
Path to the lab
While drawn to science from an early age, Lichtman’s interests in school were never confined to the laboratory. As an undergraduate at Bowdoin College, he started out with hopes of majoring in English and met his wife while serving as Editor-in-Chief of the school’s literary magazine. He was also for some time a political activist, growing up in the tumultuous era of the Vietnam War and beginning college in 1969, right when the U.S. was struggling to come to grips with Martin Luther King and Robert Kennedy’s assassinations the year before.
In fact, Bowdoin had to shut down for a semester in 1970 due to the degree of unrest in the country, and Lichtman was elected head of a student group that travelled to Yale for a large multi-college war protest. While acknowledging what a scary time it was, Lichtman mentions that he was taking organic chemistry that semester and jokes that he was relieved to hear the course would be graded on a pass/fail basis.
But political activism was not his cup of tea in the long term. Lichtman ended up majoring in biology, and when he was trying to decide what to do after college, several people suggested he apply to MD/PhD programs (at the time a relative novelty). The one he landed in, at Washington University School of Medicine, was and continues to be highly regarded. But Lichtman insists it was merely good fortune that brought him there. “St. Louis was so exotic from someone from the East Coast,” he says. “I didn’t know how good it was.” He also notes what an excellent deal it was at the time financially—he received an annual stipend of $2,400.
While initially finding the memorization part of medical school challenging and a bit off-putting, as soon as he was on the wards with real patients in front of him, Lichtman came to love medicine just as much as the bench research he did with Purves. “The context of learning in a natural setting made things that were so hard to memorize trivial,” he says, and adds that working with people gives you a “sense of what really matters.”
It was mainly the “primitive stage” that psychiatry was at in comparison to other medical fields that influenced Lichtman to choose a postdoctoral fellowship over residency when he completed the MD PhD program. He did basic neurobiology research on spinal cord circuitry at Harvard Medical School with Eric Frank, who is now at Tufts University. And though it is not the current focus of his lab, Lichtman has ever since maintained a strong interest in spinal cord and nerve regeneration.
Talking and teaching
In his usual humble style Lichtman likes to say that he was no good as a potential English major. But it is evident that he has deep interest and talent in communicating science. Lichtman is regularly invited for talks all over the world. Many would agree that no matter how many times he’s shared a particular research story, each time he delivers it, Lichtman does so with the passion of someone speaking on the topic for the first time, and each time he starts right from the beginning of the story, making sure his audience is on-board.
“I was very impressed by the humility and wisdom that I felt emanated from Dr. Lichtman,” wrote one high school teacher who heard him speak at a summer workshop on an anonymous feedback form. “I found his work to be singularly exciting.”
“I wish I could take his courses,” lamented another teacher, envious of the students at Harvard.
In addition to teaching neurobiology courses for undergraduates, Lichtman co-directs a semester-long graduate course called “Talking about Science.” He’s also lectured for the Knight Science Journalism Program at MIT. But perhaps the best evidence of his ability to inspire good science communication is the fact that Lichtman’s daughter is prominent science journalist Flora Lichtman—known to many through her years on NPR’s “Science Friday” series, now doing work for the NY Times and NPR Morning Edition in addition to creating biology videos for the Howard Hughes Medical Institute.
“I like talking to young people,” Lichtman says, describing part of his teaching duties as opening students’ eyes to “the wonders of asking questions that you can’t find answers for on Wikipedia.”
In addition to talking and teaching science to crowds of all sorts, Lichtman and his lab members have recently been experimenting with crowdsourcing their data analysis. Much of the image reconstruction work required to the map the connectome is labor-intensive, yet something one can learn to contribute to with a few days of training, even at the high school level—hence it’s a project amenable to being parceled out in small pieces to hundreds or even thousands of neural circuit “tracers.”
Which is why, on any given day if you go into Lichtman’s lab, you’re likely to see students of varying ages, from varying parts of US and even abroad, scanning through colorful images of brain slices on computers and carefully marking the boundaries of cellular structures. Some groups of students have even traced down to the level of individual synaptic vesicles, of which there may be several hundred at a single synaptic site.
Embracing complexity
Lichtman can give you a lot of reasons to be anxious about the state of science in America today. He’s deeply concerned about the lack of funding and interest in basic research, and believes these problems may be graver now than ever before in his lifetime.
But the trend towards “big data”— the unbiased, large scale datasets gathered for connectomics, genomics, proteomics, informatics, and countless other fields—is one he finds quite reassuring. Calling it a “watershed type moment, a really special time in neurobiology and other biologies,” Lichtman says we’ve reached a time when scientists recognize a need to prioritize gathering data over testing preconceived hypotheses.
“We are awed by the majesty and complexity of the world,” Lichtman observes. “You can’t put New York City or Boston into a word or a sentence. There’s just too much stuff going on…. We just have to start thinking about biology the same way. The same wonder we feel about a city we can feel about a cell or a brain.”
This instinct to embrace complexity and observe without manipulation is apparent even when Lichtman reminisces about the pond water experiments of his childhood. He is quick to point out, for instance, that the refined cultures of microorganisms such as paramecia given to students in grade school biology classes, while interesting, are nowhere near as rich or teeming with diversity as natural pond water. This is no different from his philosophy on the danger of cherry-picking specific neurons or molecules to focus on when mapping brain circuits, and why he’s worked so hard on the Brainbow and electron microscopy technology development.
He likens the concepts of information and understanding in biology to two ends of a seesaw. The more information you have, the less you understand. And if you claim to understand a lot, it’s because you have too little information.
This means a scientist’s work is never finished. It’s been thirty years since Lichtman started his lab. Yet the most important research question is always the one he’s working on currently and the hardest time is always the present, he says. “I feel very much like the big challenge is ahead of me.”